Hydrogen Deuterium Exchange
From LEAP
(→The Process:) |
(→What does the H/D-X PAL do?) |
||
| Line 67: | Line 67: | ||
'''Quenching''' The labeling reaction is stopped by transferring a volume of the mixture to an excess of quenching reagent in a pre-cooled vessel. | '''Quenching''' The labeling reaction is stopped by transferring a volume of the mixture to an excess of quenching reagent in a pre-cooled vessel. | ||
| - | '''Injection''' A measured portion of the quenched reactants is injected into a multi-column, multi-valve LCMS system. This step is synchronized with the analytical system using hardware signals. The PAL will inject samples as the LCMS system becomes ready. Labeled and quenched reactants are held in the cooled stack until they have been injected and analyzed. | + | '''Injection''' A measured portion of the quenched reactants is injected into a multi-column, multi-valve LCMS system. This step is synchronized with the analytical system using hardware signals. The PAL will inject samples as the LCMS system becomes ready. Labeled and quenched reactants are held in the cooled stack until they have been injected and analyzed. |
| - | + | ||
=== Software Control === | === Software Control === | ||
Revision as of 17:38, 14 April 2009

| Hydrogen Deuterium Exchange |
| Application Type | |
| SPECIAL | |
| Application ID | |
| HD-X PAL | |
| Description | |
| Automated Sample Prep for HD-X |
Contents |
General Description
Hydrogen Deuterium Exchange (HD-x) is an experimental technique to obtain structural data on proteins. It relies on the accurate measurement of the degree of labeling of the protein by deuterated hydrogen during a precisely measured labeling interval. The labeling reaction is stopped by addition of a quenching reagent, and loss of label between the quench step and analysis (usually by LCMS) is minimized by keeping the reactants at a temperature close to zero degrees Celsius.
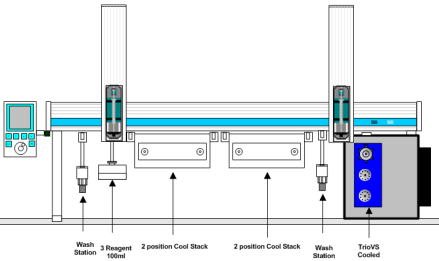
The technique is difficult to perform manually due to the demands for precise timing and reproducing exactly the same experimental conditions. Automation solves both of these challenges. LEAP has developed a robotic solution to the sample Prep and introduction to the LCMS. It consists of a Twin PAL workstation configured as shown below. There are two cooled zones on the workstation: A cooled stack at 1oC for the samples and labeled reactants and a cooled chromatography module which contains the injection valve, the column selection valves and the columns themselves. These are maintained also at around 1degree C. The automation process is controlled using LEAP Shell software.
The Process:
This technique relies on accurate timing of the labeling step, and in maintaining low temperatures for all steps after the labeling reaction has been stopped (quenched). Most experiments require labeling times in a range from 10 seconds to 8 hours. Precision in the timing is important, especially for the shorter times. It is important to schedule the setup of the labeling reactions so as to avoid timing conflicts when reactions must be stopped and reactants injected to the chromatography system. One of the biggest challenges to manually performing this technique is the correct timing of each step. LEAP has developed a scheduling application using their proprietary software “LEAP Shell” which will automate the labeling of samples, as well as the injection to the MS and the associated chromatography.
A typical experiment would consist of around a dozen time points for each protein sample, ranging from 10 seconds to 8 hours. The PAL system will schedule the experiments and perform each of the following 3 steps for each time point and sample:
- Labeling The protein is mixed with a Deuterium reagent and incubated for a precisely timed interval at room temperature.
- Quenching The labeling reaction is stopped by transferring a volume of the mixture to an excess of quenching reagent in a pre-cooled vessel (0oC).
- Injection A measured portion of the quenched reactants is injected into a multi-column, multi-valve LCMS system. This step is synchronized with the analytical system using hardware signals. The PAL will inject samples as the LCMS system becomes ready. Labeled and quenched reactants are held in the cooled stack until they have been injected and analyzed.
Chromatography consists of a digestion stage as sample is passed through an enzymatic column. Digested fragments are collected onto a trap column. After this step, they are eluted to the analytical column and to the MS. The Chromatographic runtime is around 15 minutes. When starting a run, the program first checks which samples are to be set up early (by default the threshold for this is 1 hour labeling time or longer). It sets these up and starts the labeling timers for each one. It also checks which samples have rapid label time (by default less than 60 seconds). These must be labeled and quenched in a single step to achieve maximum speed. Minimum label times are around 10 seconds. Key features of the Hydrogen Deuterium scheduler application:
- No need for the operator to worry about timing conflicts for the labeling steps. These are resolved by the program on the fly.
- Easy entry of the labeling experiments in ascending order of labeling time.
- No limit to the number of protein samples which can be handled in a single experiment.
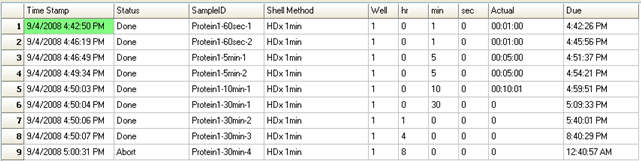
- Run-time display of the measured labeling time next to the requested labeling time (accuracy is within 1 second).
- Run-time display of the status of each sample as it progresses through the experiment e.g. “Labeling”, “quenched”, “injected”.
- The speed of the PAL is monitored by the program and timing adjustments are made on the fly to maintain incubation accuracy e.g. No need to worry about timing if PAL wash parameters change.
- Logfile output of all experimental conditions and the actual labeling and injection times of each sample.
- Long labeling time experiments are automatically set up first, to get the process started as soon as possible.
- Samples are injected as the LCMS system comes ready. LC runtime does not impact the efficiency of the labeling experiments.
- Can be integrated with the following data systems: Analyst 1.4.2, Xcalibur, ChemStation.
Significant Markets
- Pharmaceutical - Drug Discovery,
- Academia - Protein structural studies
Protein Structure
Protein function is strongly related to their tertiary structure and conformation. Chemical interactions which change this conformation can significantly affect protein functionality. This is important in studying disease states or effectiveness of drug candidates.
There are 3 commonly used techniques to study protein structure:
1. Crystallography
2. NMR
3. HD Exchange
HD Exchange is a good approach in cases where there is insufficient sample (NMR) or the protein will not crystallize. It relies on the accurate measurement of the degree of labeling of a protein by deuterated hydrogen during a precisely measured labeling interval. The deuterium is exchanged with hydrogen atoms on the exposed amide backbone of the protein and depending on the which regions are exposed and how the protein is folded, differences are seen in the MS spectra of the digested protein when compared with non-labeled proteins.
Labeling time is important (exposure time of the protein to Deuterium) as is prevention of "back-exchange" after the labeling has occurred. This is the natural tendency of normal hydrogen to replace the deuterium after the label is removed. This is minimized by reducing the temperature of the mixture to close to zero degrees Celsius.
What is an HD-X experiment ?
Performing HD-X experiments manually involves setting up many labeling mixtures then quenching these mixtures at precisely timed intervals (by changing the pH) and then injecting them onto a chilled injection valve to the LCMS. In most cases a range of label times is used, from 10 seconds to several hours. Sources of error (apart from normal liquid handling inaccuracies) are timing errors, and degradation of sample due to "back-exchange". All of these sources of error are eliminated using the automated system described here.
What does the H/D-X PAL do?
The instrument has two cooled zones for sample storage and labeling. The injection valve is chilled to 1 degree Celsius and includes a solvent pre-chiller and column selection valve for in-line protein digestion.
A typical experiment would consist of around a dozen time points for each protein sample, ranging from 10 seconds to 8 hours. The PAL system will schedule the experiments and perform each of the following 3 steps for each time point and sample:
Labeling The protein is mixed with a Deuterium reagent and incubated for a precisely timed interval at room temperature.
Quenching The labeling reaction is stopped by transferring a volume of the mixture to an excess of quenching reagent in a pre-cooled vessel.
Injection A measured portion of the quenched reactants is injected into a multi-column, multi-valve LCMS system. This step is synchronized with the analytical system using hardware signals. The PAL will inject samples as the LCMS system becomes ready. Labeled and quenched reactants are held in the cooled stack until they have been injected and analyzed.
Software Control
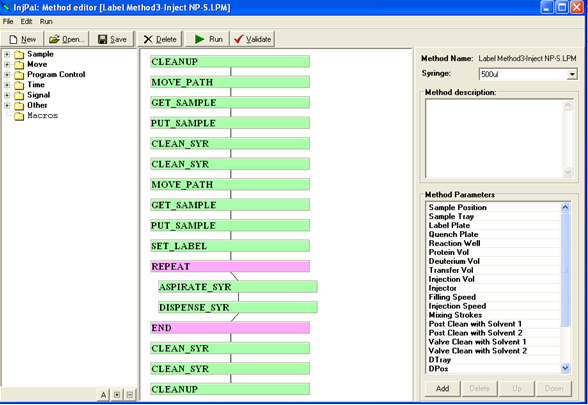
LEAP has developed a scheduling application using their proprietary software “LEAP Shell” which will automate the above labeling steps, as well as the injection to the MS and the associated chromatography.
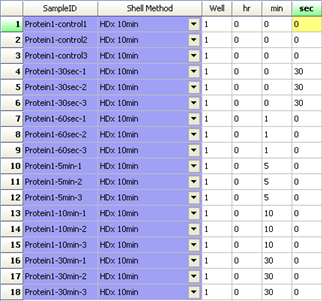
LEAP Shell Screen Shot 1: User interface to set up the labeling experiment:
LEAP Shell Screen Shot 2: Powerful editor for the PAL Methods:
LEAP Shell Screen Shot 3: Run-time display of the labeling experiment with actual times:
Useful links
HXMS.com A good general reference site on HD Exchange theory and practice [1]
ASMS Interest Group - Hydrogen Exchange and Covalent Labeling [2]
LEAP Website H/D-x product description [3]
Publications